1) Simulate data
under an evolutionary scenario with parameter
values defined in an input parameter file
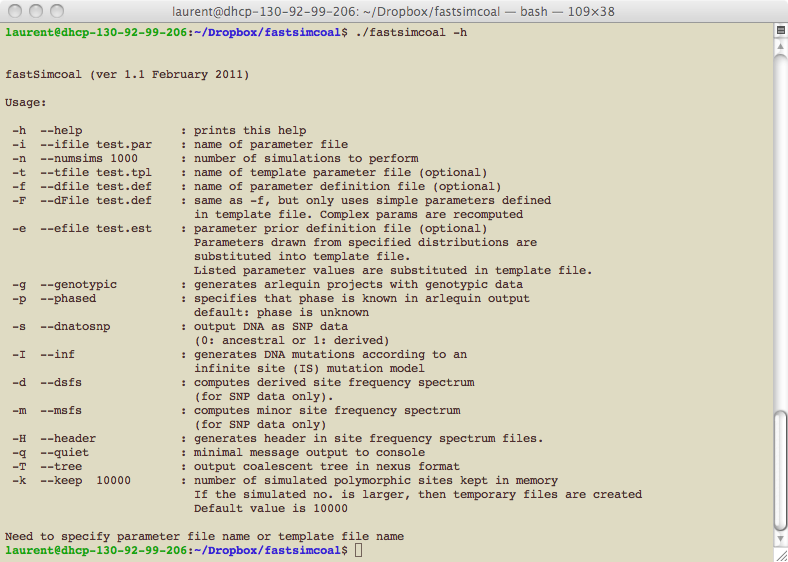
./fastsimcoal
-i test.par -n 100
fastsimcoal will use the scenario and the
parameter values defined in the parameter file test.par
and make 100 simulations under this scenario.
2) Simulate data under an evolutionary scenario
with parameter
values randomly drawn from priors
./fastsimcoal –t test.tpl –n 10 –e test.est –E 100
fastsimcoal will use the scenario defined
in the template file test.tpl and generate 100 sets of
parameter values by randomly drawing these values from the priors
defined in
the file test.est. 10 simulations
will be done for each sets of randomly drawn parameter values.
3) Simulate data under an evolutionary scenario
with parameter
values defined in an external definition file
./fastsimcoal -t test.tpl -n 100 -f test.def
fastsimcoal
will use the scenario defined in the template file test.tpl and use the
parameter values found in the definition file test.def.
100 simulations will be done for each set of predefined
parameter values.